1. Why EC number?
The basis of linking genomics and chemistry is the EC numbers [JACS, 2004; Enzyme Nomenclature, 2002; Bioinformatics, 2009; PLOS Comp. Bio. 2010]. The EC numbers represent enzymatic reactions (chemical information), but they are also utilized as identifiers of enzymes and enzyme genes (genomic information). The EC number plays a key role in classifying enzymatic reactions and in linking the enzyme genes or proteins to reactions in metabolic pathways. This duality of the EC numbers makes it possible to link the genomic repertoires of enzyme genes to the chemical repertoire of metabolic pathways.2. Why EC Assignment?
The assignment of the EC numbers is performed manually, based on published experimental data on individual enzymes, by the Joint Commission on Biochemical Nomenclature of the International Union of Biochemistry and Molecular Biology and the International Union of Pure and Applied Chemistry.Unfortunately, there are numerous reactions known to be present in various pathways, but they will never get EC numbers because the EC number assignment requires published articles on full characterization of enzymes [JACS 2004]. There are numerous reactions known to be present in various pathways but without any official EC numbers, most of which have not hope to be given ones because of the lack of the published articles on enzyme assays [Bioinformatics, 2009].3. Former Studies on EC assignments?
There are some systems purely based on chemical knowledge, without any use of protein sequence or other information on enzymes [JACS, 2004; Bioinformatics 2009; JCIM 2009; Bioinformatics, 2008; PLOS Comp. Bio. 2010]. The published methods are mainly developed by (1) reactant pairs and RDM patterns; (2) physico-chemical descriptors; and (3) atom and bond difference.4. Why need a new tool to assign EC numbers?
E-zyme will need manually defined RDM patterns to assign EC numbers for whole reactions, although E-zyme can automatically assign EC numbers for two molecules. MolMap is based on a commercial package (Petra) to assign EC numbers for reactions. A method purely using molecular structure is very useful and feasible in applications. PLOS Comp. Bio. 2010 applied atom and bond difference to assign EC numbers. Both MolMap and PLOS Comp. Bio. 2010 have not web server.5. What is ECAssigner?
ECAssigner is a tool to assign EC numbers for input enzymatic reactions using reaction difference fingerprints and euclidean distance. ECAssigner is based on reaction similarity described by difference fragments of molecular structures.6. Why reaction similarity and reaction difference fingerprints?
The automatic perception of similarities between metabolic reactions, i.e. their classification, is a chemoinformatics issue with an impact in bioinformatics, biotechnology, or medicinal chemistry [Diogo Sousa, JCIM 2009]. There are some researchers developed different methods to measure reaction similarity between individual steps of enzymatic reaction mechanisms and to quantitatively measure the similarity of enzymatic reactions based upon their explicit mechnisms [O'Boyle, N.M.et al. Using reaction mechanism to measure enzyme similarity. J. Mol. Biol. 2007, 368, 1484-1499. ]. Ridder and Wagener clustered a data set of metabolic reactions using a difference fingerprint defined by te differences in occurence of each Syble atom type in the reactant and product fingerprints [Ridder, L.; Wagener, M. SyGMa: Combining expert knowledge and empirical scoring in the prediction of metabolites. ChemMedChem. 2008,3,821-832.]. Faulon et al [20] employed molecular signatures of topological atom neighborhoods to derive reaction signatures and used such descriptors with SVM to classify metabolic reactions in terms of EC numbers[Faulon et al. Bioinformatics, 2008, 24, 2335-2349.]. In RxnFinder, our group has successfully applied reaction similarity to search biochemical reactions [Bioinformatics 2011]. In our studies, reaction difference fingerprints, which not only consider atom types, but also bond connections, are applied in assigning EC numbers.7. How ECAssigner works?
(1) The training reaction data used in ECAssigner is selected from 8105 KEGG reactions after excluding those reactions with missing molecules (unbalanced reactions), without Glycans, with EC assignments, without molecules having not structure information, and/or without polymer-like molecules, which are shown along with each KEGG reaction ID;
(2) Each molecule of a reaction will be calculated to get a list of molecular fragments using a fragmentation method ;
(3) The molecular fragments of reactant molecules minus the fragments of product molecules will result in the reaction difference fingerprints ;
(4) reaction difference fingerprints will be computed for both the input reactions from users and the training reactions;
(5) Euclidean distance will be calculated for the input reactions and the training reactions;
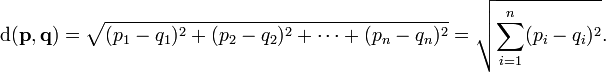
8. EC assignment examples?
Examples (R00005-R00131):R00005 (urea-1-carboxylate amidohydrolase) Urea-1-carboxylate + H2O <=> 2 CO2 + 2 NH3 (C01010 + C00001 <=> 2 C00011 + 2 C00014): AssignedEC: ['3.5.1']; Entry R00005 Reaction Name urea-1-carboxylate amidohydrolase Definition Urea-1-carboxylate + H2O <=> 2 CO2 + 2 NH3 Equation C01010 + C00001 <=> 2 C00011 + 2 C00014
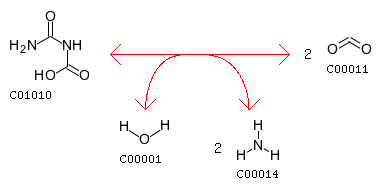
The closest reaction (with the Minimum distance: 18): R00131 Entry R00131 Reaction Name Urea amidohydrolase Definition Urea + H2O <=> CO2 + 2 NH3 Equation C00086 + C00001 <=> C00011 + 2 C00014
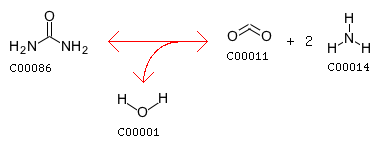
Reaction difference fragments: {'O.co2': -2, 'C1=O.co2': -2, 'Nam@C2@Nam': 1, 'C2@Nam': 2, 'Nam': 2, 'O.co2=C1=O.co2': -1, 'Nam@C2=O2': 2, 'C2': 1, 'C2=O2': 1, 'C1': -1, 'N3': -2, 'O3': 1, 'O2': 1}
Examples (R00008-R00237):
R00008 with a AssignedEC: ['4.1.3']; Entry R00008 Reaction Name 4-hydroxy-4-methyl-2-oxoglutarate pyruvate-lyase (pyruvate-forming) Definition Parapyruvate <=> 2 Pyruvate Equation C06033 <=> 2 C00022
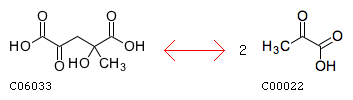
The closest reaction (Minimum distance: 2): rxnID: R00237 with a AssignedEC: ['4.1.3']; Entry R00237 Reaction Name (3S)-citramalyl-CoA pyruvate-lyase (acetyl-CoA-forming) Definition (3S)-Citramalyl-CoA <=> Acetyl-CoA + Pyruvate Equation C01011 <=> C00024 + C00022
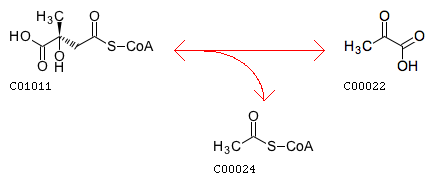
Reaction difference fragments: {'C3~C2~Cac~O.co2': -1, 'C3~C3~Cac~O.co2': 2, 'C3~C2=O2': -1, 'O.co2~Cac~C2=O2': -1, 'O.co2~Cac~C3~O3': 1, 'C3~C3~Cac': 2, 'O.co2=Cac~C3~O3': 1, 'O3': 1, 'C2~C3~C3~Cac': 1, 'O2': -1, 'C3~C3~Cac=O.co2': 2, 'C3~Cac~O.co2': 1, 'C3': 1, 'C2': -1, 'C2~C3': -1, 'C2~C3~C3~C3': 1, 'C3~C3~C2~S3': 1, 'C3~O3': 1, 'C2~Cac=O.co2': -1, 'Cac~C3~O3': 1, 'Cac~C2=O2': -1, 'C3~Cac': 1, 'C3~C3~C2=O2': 1, 'C3~C3~O3': 2, 'C3~C2~Cac=O.co2': -1, 'O.co2=Cac~C2=O2': -1, 'C3~Cac=O.co2': 1, 'C2~Cac': -1, 'C2~C3~C3~O3': 1, 'C3~C3': 2, 'C3~C3~C3': 1, 'C2~Cac~O.co2': -1, 'C2=O2': -1, 'C3~C2~Cac': -1, 'C2~C3~C3': 1}
9. Path length selection in ECAssigner?
In the model development, different fragment lengths are used to get cross-validation prediction.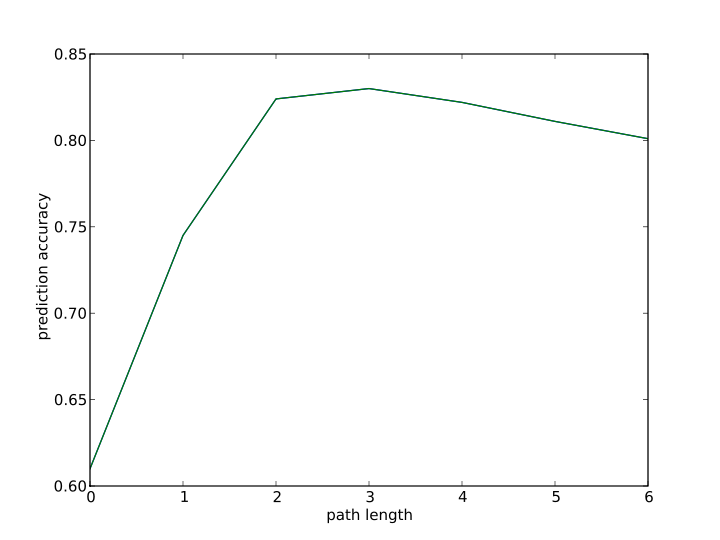
10. Performance of selected ECAssigner model?
The prediction accuracy is 0.830; 0.865, 0.922 for sub-subclass, subclass, main class respectively. On different main classes. From the table, EC2>EC3>EC1>EC6>EC4>EC5. 82.6% reactions are assigned EC numbers on main classes EC1,2,3. Reasons on why EC4,5,6 are very low?11. Number of reactions predicted along with the distance changes?
In the model development, the EC number is assigned according to the Euclidean distance of the input reaction to the training reactions. The prediction reactions are counted with the distance changes as shown in the distance-count plot.From the table, 84.6% reactions are predicted with distance smaller than 20, in which 89.4% predictions are correct. From the results, the more similar (smaller distance), the higher accuracy.
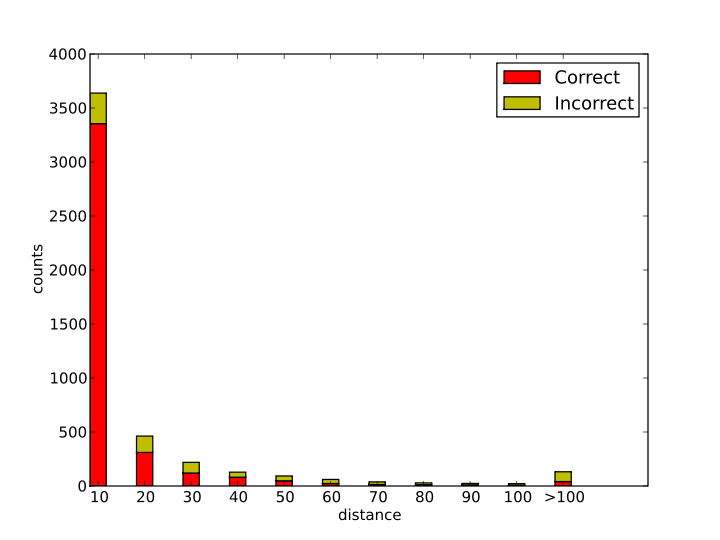
From the plot, more than 84.6% of the reactions are predicted with distance smaller than 20. Some reactions are very disimilar with the existing reactions.
rxnID: R07251 with a AssignedEC: ['2.3.1']; Entry R07251 Reaction Name acyl-CoA:malonyl-CoA C-acyltransferase (decarboxylating, oxoacyl- and enoyl-reducing, thioester-hydrolysing) Definition Acetyl-CoA + 8 Malonyl-CoA + 11 NADPH + 11 H+ + S-Adenosyl-L-methionine <=> Dihydromonacolin L + 9 CoA + 8 CO2 + 11 NADP+ + S-Adenosyl-L-homocysteine + 6 H2O Equation C00024 + 8 C00083 + 11 C00005 + 11 C00080 + C00019 <=> C15536 + 9 C00010 + 8 C00011 + 11 C00006 + C00021 + 6 C00001
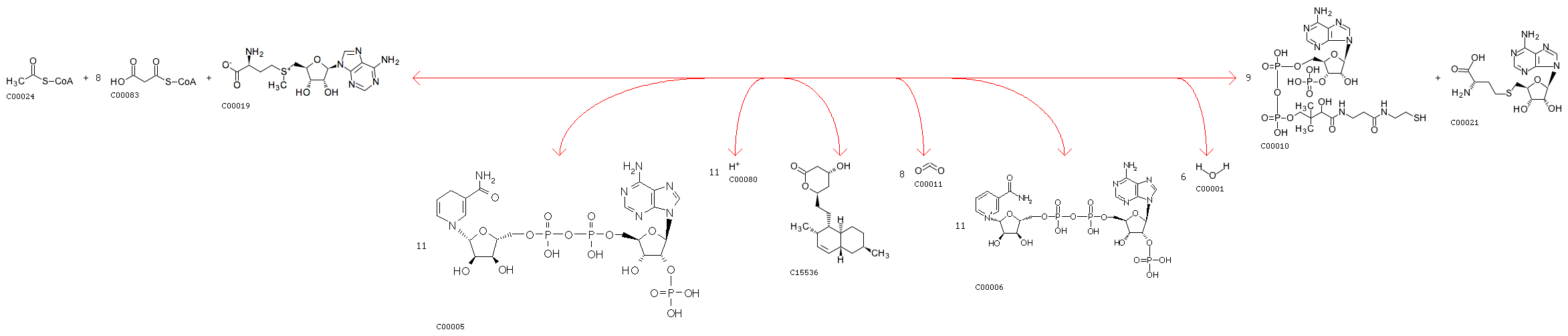
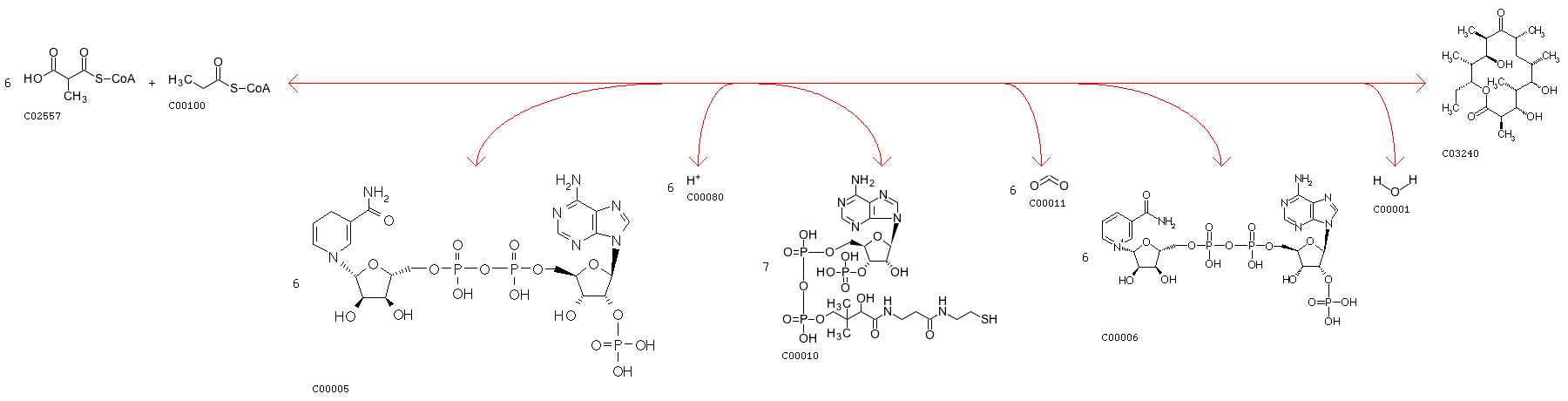
Minimum distance: 6078; rxns Refered: ['R00918']; rxnID: R00918 with a AssignedEC: ['2.3.1']; Entry R00918 Reaction Name propanoyl-CoA:methylmalonyl-CoA malonyltransferase (cyclizing); propanoyl-CoA:methylmalonyl-CoA 2-C-acyltransferase (decarboxylating, oxoacyl-reducing and cyclizing); Malonyl-CoA:propionyl-CoA malonyltransfersase (cyclizing) Definition 6 Methylmalonyl-CoA + Propanoyl-CoA + 6 NADPH + 6 H+ <=> 7 CoA + 6-Deoxyerythronolide B + 6 CO2 + 6 NADP+ + H2O Equation 6 C02557 + C00100 + 6 C00005 + 6 C00080 <=> 7 C00010 + C03240 + 6 C00011 + 6 C00006 + C00001
12. Prediction accuray with the distance changes?
In the model development, the EC number is assigned according to the Euclidean distance of the input reaction to the training reactions. The prediction accuracy is calculated with the distance changes as shown in the distance-accuracy plot.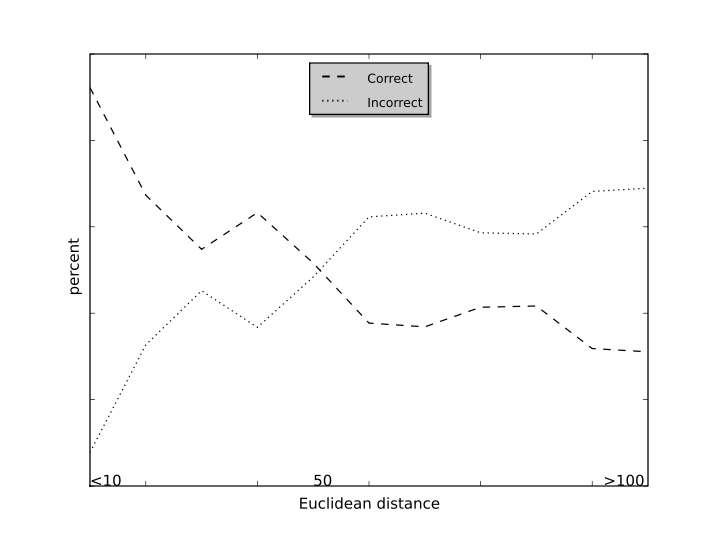
From the cross-validation results, the prediction based on distance smaller than 50 will lead to high accuracy.
13. Incorrect predictions?
There are totally 824 incorrect predictions. (All incorrect reactions are list 824 Incorrect Predictions ) Some reactions are checked to find the reasons.Examples (R00004-R00764):
R00004 Entry R00004 Reaction Name diphosphate phosphohydrolase; pyrophosphate phosphohydrolase Definition Diphosphate + H2O <=> 2 Orthophosphate Equation C00013 + C00001 <=> 2 C00009

The closest reaction: R00764 Entry R00764 Reaction Name diphosphate:D-fructose-6-phosphate 1-phosphotransferase Definition Diphosphate + D-Fructose 6-phosphate <=> Orthophosphate + D-Fructose 1,6-bisphosphate Equation C00013 + C00085 <=> C00009 + C00354
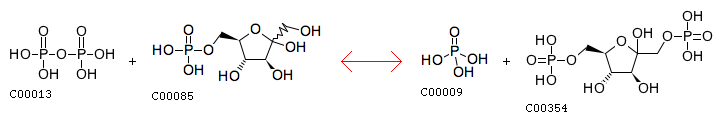
Examples (R00012-R04509):
R00012 Entry R00012 Reaction Name GTP:GTP guanylyltransferase Definition 2 GTP <=> Diphosphate + P1,P4-Bis(5'-guanosyl) tetraphosphate Equation 2 C00044 <=> C00013 + C01261

The closest reaction: Entry R04509 Reaction Name ATP:4-amino-2-methyl-5-phosphomethylpyrimidine phosphotransferase Definition ATP + 4-Amino-2-methyl-5-phosphomethylpyrimidine <=> ADP + 2-Methyl-4-amino-5-hydroxymethylpyrimidine diphosphate Equation C00002 + C04556 <=> C00008 + C04752

Examples (R00016-R01057):
R00016: Entry R00016 Reaction Name D-glucose-1-phosphate:D-glucose-1-phosphate 6-phosphotransferase Definition 2 D-Glucose 1-phosphate <=> D-Glucose + D-Glucose 1,6-bisphosphate Equation 2 C00103 <=> C00031 + C00660
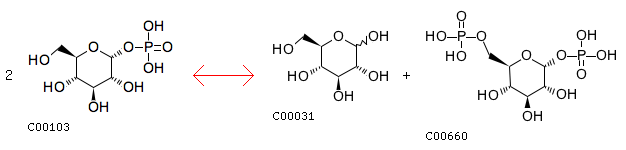
The cloest reaction: R01057 Entry R01057 Reaction Name D-Ribose 1,5-phosphomutase Definition alpha-D-Ribose 1-phosphate <=> D-Ribose 5-phosphate Equation C00620 <=> C00117 Comment intermediate (see [CPD:C01151])

numEC1Diff 378 numEC2Diff 654 numEC3Diff 824
14. Comparisons with other EC prediction methods?
Comparisons with E-zyme: Comparisons with MOLMAP method:| Method | Method Basis | Automatic for Whole Reaction | Online Server |
|---|---|---|---|
| E-zyme | RPAIR (manual) | No | Yes for two molecules |
| MolMap | Petra package (commercial) | Yes | No |
| Egelhofer et al. | Atom and Bond difference | Yes | No |
| ECAssigner | Reaction Difference Fingerprints with Variable Length | Yes | Yes |