Q: What is VNP:
A: VNP is developed to Visualize Network Pharmacology of targets, diseases, and drugs with a graph network by using disease, target or drug names, chemical structures or protein sequence. Currently, there are more than 1,000 diseases, more than 500 protein targets, and more than 1,500 drugs crated with tens of thousands of connections among them.
Q: Functions provided in VNP?
A: VNP addressed applications of interactive visual network pharmacology tools to network
<1> diseases, <2> targets and <3> drugs in a dynamics web-based network graph.
<1> diseases, <2> targets and <3> drugs in a dynamics web-based network graph.
Q: How to use VNP?
A: VNP mainly contains three functional modules: drug-centric, target-centric and disease-centric visual network pharmacology. By input a keywords, VNP will generate an online interactive network (diseases-targets-durgs) view of the retrieved records.
1. For a basic searching: Users can search database using disease name "Disease-Centric Network Pharmacology", target name "Target-Centric Network Pharmacology" or drug name "Drug-Centric Network Pharmacology" which they are interesting in to obtain a network view.
2.Further, users can search database using drug substructures (smiles need) "Drug-Centric Network Pharmacology Using Molecular Sub-Structure" to get a Drug-Centric Network view or using protein sequence similarity (fasta format need) "Target-Centric Network Pharmacology Using Protein Sequence" to get a Target-Centric Network view.
3. In the obtained network view, each node is a disease, target or drug, and each edge is a known connection between two of them.
4. Users can only search one kind of keywords one time.
5. When you move mouse on any node, relevant information can be seen.
6. Click on any node on the graph, users will get a detailed list of the related records.
1. For a basic searching: Users can search database using disease name "Disease-Centric Network Pharmacology", target name "Target-Centric Network Pharmacology" or drug name "Drug-Centric Network Pharmacology" which they are interesting in to obtain a network view.

Search Drug via the example
2.Further, users can search database using drug substructures (smiles need) "Drug-Centric Network Pharmacology Using Molecular Sub-Structure" to get a Drug-Centric Network view or using protein sequence similarity (fasta format need) "Target-Centric Network Pharmacology Using Protein Sequence" to get a Target-Centric Network view.
3. In the obtained network view, each node is a disease, target or drug, and each edge is a known connection between two of them.
4. Users can only search one kind of keywords one time.
5. When you move mouse on any node, relevant information can be seen.
6. Click on any node on the graph, users will get a detailed list of the related records.
Q: Searching options provided in VNP?
A: VNP search options are disease, target or drug keywords, drug structure, and protein sequence.
Q: Searching cases available in VNP?
A: Three search examples are illustrated:
1. A disease-centric network retrieved by Alzheimer's disease: Input disease string as "Alzheimer's disease" (Figure 1) and click the search button following the input text area. The graph (Figurec2) provides an intuitive view to network the targets and drugs related with Alzheimer's disease. The connection degrees between targets and drugs are asymmetry, for instance, 3 drugs connecting to acetylcholinesterase. One drug is linked to Alpha-2 adrenergic receptor.
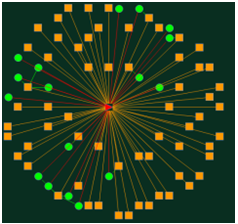
2. A target-centric network obtained by Muscarinic acetylcholine receptor: Query string "Muscarinic acetylcholine receptor" (figure3) in the Target-Centric Network Pharmacology text blank, and click the search button following the input text area. It will obtain six targets (figure4). These targets are found connecting with more than twenty diseases and fifty drugs.
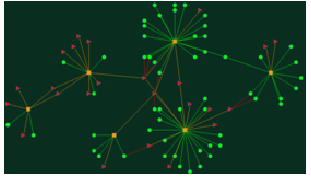
3. A drug-centric network searched by chemical substructure (7-Amino-3-cephem-4-carboxylic acid): Search substructure (7-Amino-3-cephem-4-carboxylic acid) as smiles string: "NC1C(=O)N2C1SCC=C2C(=O)O" in text blank(Figure 5) and click the search button following the input text area. We will get more than thirty drugs containing the chemical substructure. These drugs are further mapped with six diseases and six targets.
Tips: 1) Red triangles, green circles and yellow rectangles correspond to diseases, drugs, and targets respectively. 2) Click on any node on the graph, users will get a detailed list of the related records.
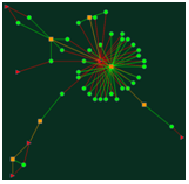
1. A disease-centric network retrieved by Alzheimer's disease: Input disease string as "Alzheimer's disease" (Figure 1) and click the search button following the input text area. The graph (Figurec2) provides an intuitive view to network the targets and drugs related with Alzheimer's disease. The connection degrees between targets and drugs are asymmetry, for instance, 3 drugs connecting to acetylcholinesterase. One drug is linked to Alpha-2 adrenergic receptor.

Figure 1

Figure 2
2. A target-centric network obtained by Muscarinic acetylcholine receptor: Query string "Muscarinic acetylcholine receptor" (figure3) in the Target-Centric Network Pharmacology text blank, and click the search button following the input text area. It will obtain six targets (figure4). These targets are found connecting with more than twenty diseases and fifty drugs.

Figure 3

Figure 4
3. A drug-centric network searched by chemical substructure (7-Amino-3-cephem-4-carboxylic acid): Search substructure (7-Amino-3-cephem-4-carboxylic acid) as smiles string: "NC1C(=O)N2C1SCC=C2C(=O)O" in text blank(Figure 5) and click the search button following the input text area. We will get more than thirty drugs containing the chemical substructure. These drugs are further mapped with six diseases and six targets.
Tips: 1) Red triangles, green circles and yellow rectangles correspond to diseases, drugs, and targets respectively. 2) Click on any node on the graph, users will get a detailed list of the related records.

Figure 5

Figure 6
Q: How searches should not be done?
A: 1. In "Drug-Centric Network Pharmacology" searching, only the keywords of drug name can be accepted. Users should not search other keywords such as: Smiles format, Chemicals codes etc.
2. In "Target-Centric Network Pharmacology" searching, only the keywords of target name can be accepted. Users should not search other keywords such as: target sequences, PBD id etc.
3. In "Disease-Centric Network Pharmacology" searching, only the keywords of disease name can be accepted. Users should not search other keywords such as: symptoms of the disease, OMIM id etc.
4. In "Drug-Centric Network Pharmacology Using Molecular Sub-Structure" searching, only the keywords of SMILES format of molecular sub-structure can be accepted. Users should not search other keywords such as: sub-structure name.
5. In "Target-Centric Network Pharmacology Using Protein Sequence" searching, only the keywords of FASTA format of the target protein can be accepted. Users should notice do not miss the header (">P48039") and do not search other keywords such as: target name.
6. When users input drug (or target, or disease) names, users should click the search button following the input text area. Please do not input keywords in one area, and click the search button in other text area.
7. Please do not input more than one kind keywords searching by one time.
2. In "Target-Centric Network Pharmacology" searching, only the keywords of target name can be accepted. Users should not search other keywords such as: target sequences, PBD id etc.
3. In "Disease-Centric Network Pharmacology" searching, only the keywords of disease name can be accepted. Users should not search other keywords such as: symptoms of the disease, OMIM id etc.
4. In "Drug-Centric Network Pharmacology Using Molecular Sub-Structure" searching, only the keywords of SMILES format of molecular sub-structure can be accepted. Users should not search other keywords such as: sub-structure name.
5. In "Target-Centric Network Pharmacology Using Protein Sequence" searching, only the keywords of FASTA format of the target protein can be accepted. Users should notice do not miss the header (">P48039") and do not search other keywords such as: target name.
6. When users input drug (or target, or disease) names, users should click the search button following the input text area. Please do not input keywords in one area, and click the search button in other text area.
7. Please do not input more than one kind keywords searching by one time.