RxnPatter (FAQ)
Q: What is RxnPatter?
A: RxnPatter is a web server for extracting chemical transformation patterns from biochemical reactions.
Q: What is a chemical transformation pattern?
A: Chemical transformation pattern is a combination of molecular fragments composed of reaction centers (bonds changed during a reaction).
Q: Examples on chemical transformation pattern?
KEGG Biochemical reactions: Oxidoreductases. Acting on the CH-OH group of donors. With NAD+ or NADP+ as acceptor
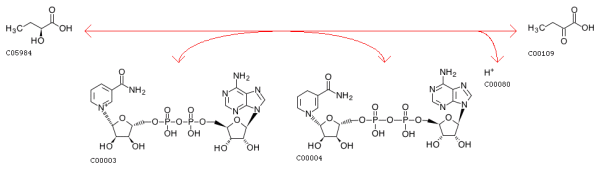
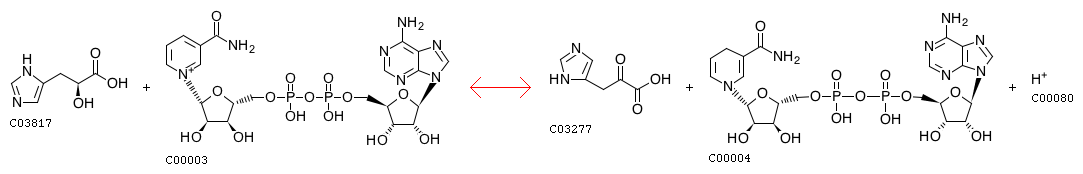
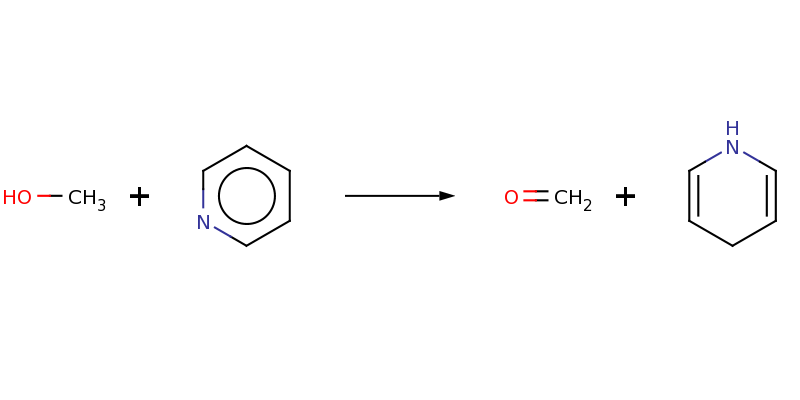
Chemical transformation pattern of the above two reactions: CO.c1cccnc1>>C=O.N1C=CCC=C1
Q: How to extract chemical transformation pattern?
A: (The first step:) Get atom alignments for a biochemical reaction. For example, atom alignments of RPAIR database from KEGG database. Atom mapping of BioPath from Gasteiger's group.
(The second Step:) Identify bonds (reaction centers) changed during a reaction. Reaction centers refer to those chemical bonds formed or broken during a reaction process.
(The third Step:) Link those bonds into a fragment for each molecule.
(The fourth Step:) Combine these fragments into a smirks string (examples are available from JCIM 1999, 39(6), 1161-1172).
Q: Where is the RxnPatter Server for extracting chemical transformation pattern?
A: RxnPatter is freely available at RxnPatter.
Q: What is the input of RxnPatter?
A: The input is a reaction smiles string (smirks) (Please refer to DayLight Documents). The input reaction must be balanced. Please refer to Balanced Reactions
Q: What is the output of RxnPatter?
A: The output is the chemical transformation pattern extracted from a biochemical reaction. The chemical transformation pattern extracted is also represented as a reaction smiles string (smirks).
Q: What is the processing methods used in RxnPatter?
A: The processing method is a recursive graph comparison method combined with an entropy function.
Q: How is RxnPatter developed?
A: The processing methods have been developed since 20008 while Prof. Qian-Nan Hu worked in the KEGG group. The RxnPatter server has been running for more than 6 months in Hu's lab. During testing, more than 1000 KEGG biochemical reactions are analyzed to validate the server. About 8 researchers from other groups have been involved in the testing.
|


